Table Of Contents
An overview of PISE
Database construction
Search single or multiple genes
Filter
IGV visualization
Data collection
Data analysis
Data analysis
References
Contact us
An overview of PISE
Welcome to Plant Splicing Efficiency Database(PISE), an online database for exploring 25,000+ Arabidopsis, 3,000+ soybean, 19,000+ rice, 17,000+ maize public RNA-Seq libraries splicing efficiency. PISE is available at https://plantintron.cn/psed/(Return to Homepage)
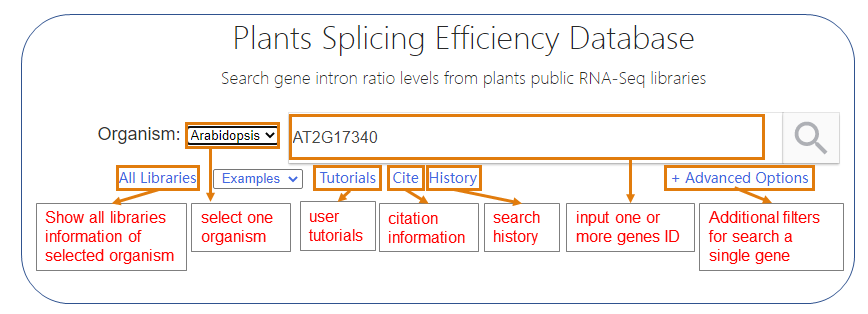 Figure 1. The overview of the databaste.
Figure 1. The overview of the databaste.
PISE is a free, web-accessible, and user-friendly database, contributes to searching, filtering, visualizing, browsing, and downloading the intron splicing efficiency data of selected organism, is described in Figure 1. PISE functions are described as follows:
● PISE collected more than 70,000 RNA-Seq libraries for Arabidopsis, maize, soybean and rice. ● PISE use a standardized pipeline to calculated the intron splicing efficiency of each gene in each library. ● PISE can search the input gene ID, and return the basic description and intron reatained levels. ● PISE supports to download and share the search results. ● PISE also has a built-in IGV-web interface.
Database construction
PISE contributes to search, visualize, browse, and download gene splicing efficiency (IRratio) data. PISE operates flexible that supports a "Google-like" search through querying of genes(Figure 1), and returns the fundamental description, exhibit and visualize IRratio levels for all intron of selected gene or on intron, and support IGV-web (https://igvteam.github.io/igv-webapp/) interfaced genomic alignment.
Search signle or multiple gene
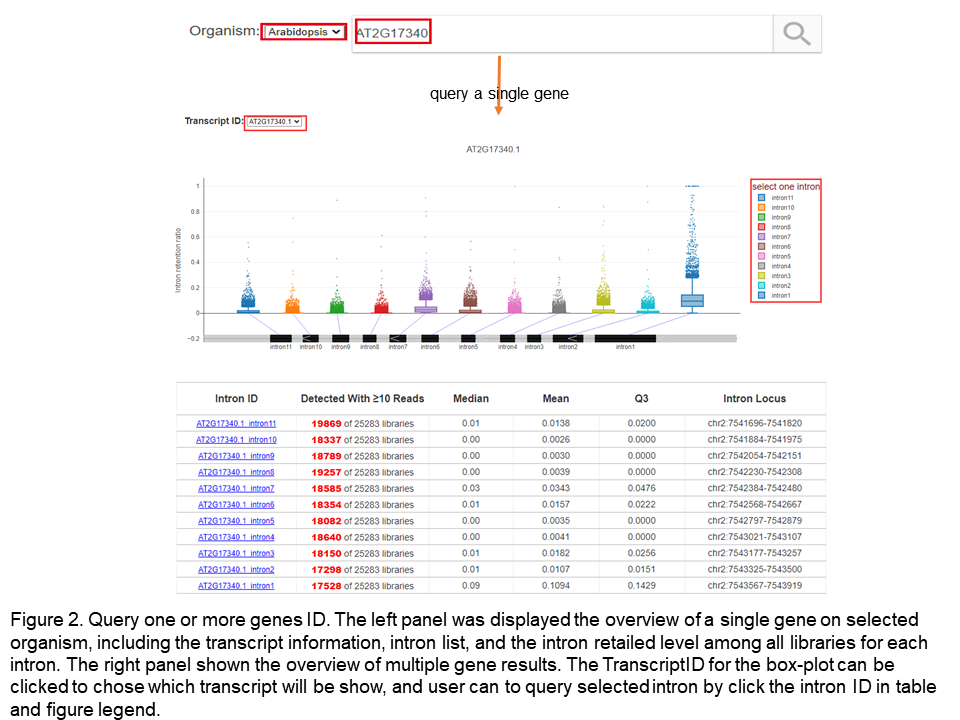
PISE allows searching four organisms one or more genes ID which at least includes one intron, including 21,908/37,336 genes annotated in Araport11 database for Arabidopsis, 31,650/39272 genes annotated in B73 v4 for maize, 43,651/55,801 genes annotated in MSU7 fro rice, 42,012/56,044 genes annotated in Williams 82 v2 for soybean (Figure 2). For visualization results, table and box plots were used to describe the global IRratio. And the table, box, and bar plots were used to display the detailed information of the selected intron among all libraries, including libraries' basic information, coverage of the intron and flank genes, the specific among different tissue, development-stage, biotic, and abiotic stress, as well as the difference of IRratio among different mutants and treatments (Figure 3).
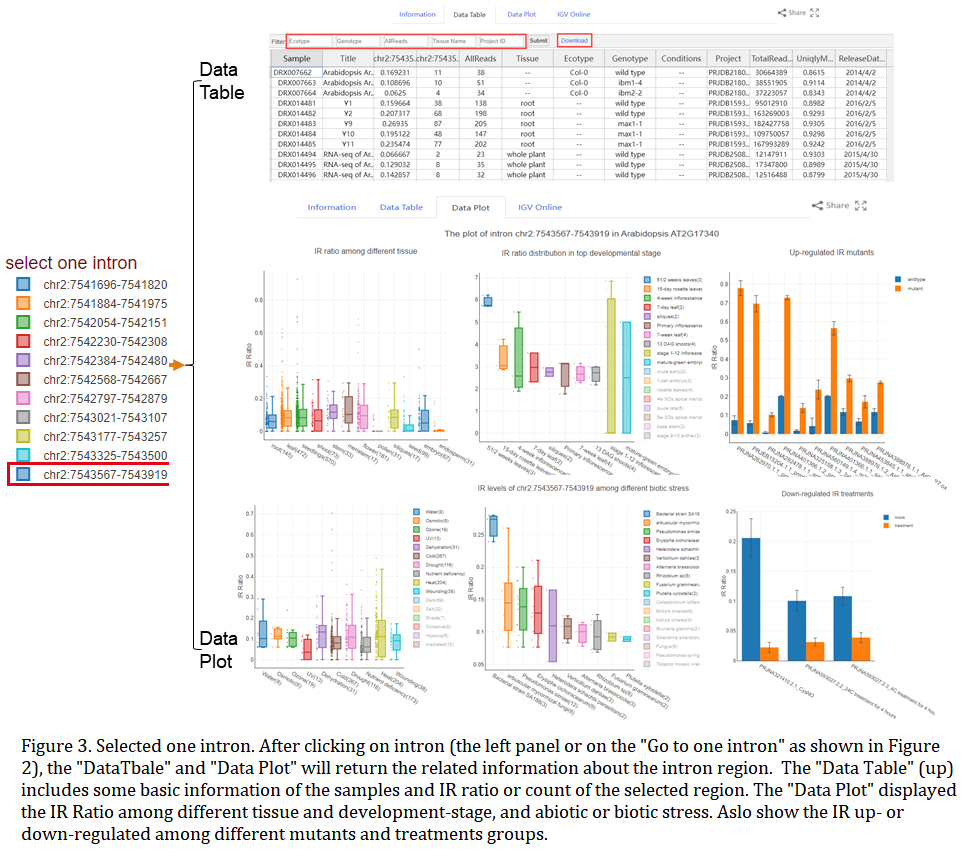
When only one gene has been searched, the advanced option will be valid to specify the maximum and minimum values of IRratio in the table. After chooseing one intron ID on the first page, the 'Data Table' will show the deatil information of the selected intron, and the "Data Plot" page will describe the highest ten groups IRratio in the development stage and stress, and the user can choose more groups to draw by clicking the legend on the right. At the same time, the page also displayed the higher IRratio levels groups of treatment conditions and mutants, which have a greater fold change of up-regulated and down-regulated, respectively. Users can download the figure or plot data by clicking the plot area button, and users also can quickly view the plot data by clicking the box or bar (Figure 3).
Filter
In the data table result, user can right-click to show advanced options for each column, such as hide, remove, order-by, filter. We also support user to filter with the submit button. For the filter option, the user can select which options to view by selecting that from their respective drop-down boxe.
IGV visualization
PISE integrates an online IGV interface to browse and compare the genome matched RNA-Seq in one library or project. The links of online IGV interface are added into the results of each type of query. If user wants to clear all current tracks, please clicking the "clear tracks" button. Also, the IGV online can add multiple libraries or projects convenient for comparison(Figure 2D).
Data collection
We use a similar strategy to collect Arabidopsis, rice, maize, and soybean RNA-Seq datasets published till 2021 from GEO, DDBJ, EBI, and SRA database. For example, the search keywords of Arabidopsis are ”((Arabidopsis thaliana[Organism]) AND "transcriptomic"[Source]) AND "RNA seq"[Strategy].” After the search, we manually went through the detailed information of all libraries and then removed pseudo-RAN-seq libraries, such as small RNA-Seq or ncRNA-Seq. Finally, we collected over 57,000 libraries with at least 1M reads aligned to the reference genome (25,283 of Arabidopsis, 10710 of rice, 3974 of soybean, and 177789 of maize).
Data analysis
Because the HISat2 supports access SRA database and can use the .sra format as input, we don't need to download raw data and convert the .sra to fastq format extra. (Because of the internet limit, some local servers can't support that function, at that time, you must use wget or other software to download raw data.) First, raw data be aligned with the reference genome by HISat2, and the alignment parameters of each organism are different. TAIR10, MSU7, Williams 82 a2, and B73 v4 were used as the reference genome for Arabidopsis, rice, soybean, and maize, respectively. Then all intron's retained values (IRratio) were calculated with python script. Finally, the DESeq2 to perform the differential analysis of the IRratio between mutants and matched controls, between treatments and matched controls.
References
Kim, D., Langmead, B., and Salzberg, S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357-360. Pertea, M., Pertea, G.M., Antonescu, C.M., et al. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290-295. Robinson, J.T., Thorvaldsdottir, H., Wenger, A.M., et al. (2017). Variant Review with the Integrative Genomics Viewer. Cancer Res 77, e31-e34. Sievert, C., Parmer, C., Hocking, T., et al.(2016). plotly: Create interactive web graphics via Plotly’s JavaScript graphing library [Software].
Contact us
Dear the PISE users, Thank you for using the PISE database! If you encounter any problem or have any question, please don't hesitate to contact us at zhaijx@sustech.edu.cn or zhangh9@mail.sustech.edu.cn. Best wishes, PISE Team